About/Help
-
What information is available in the MOI?
-
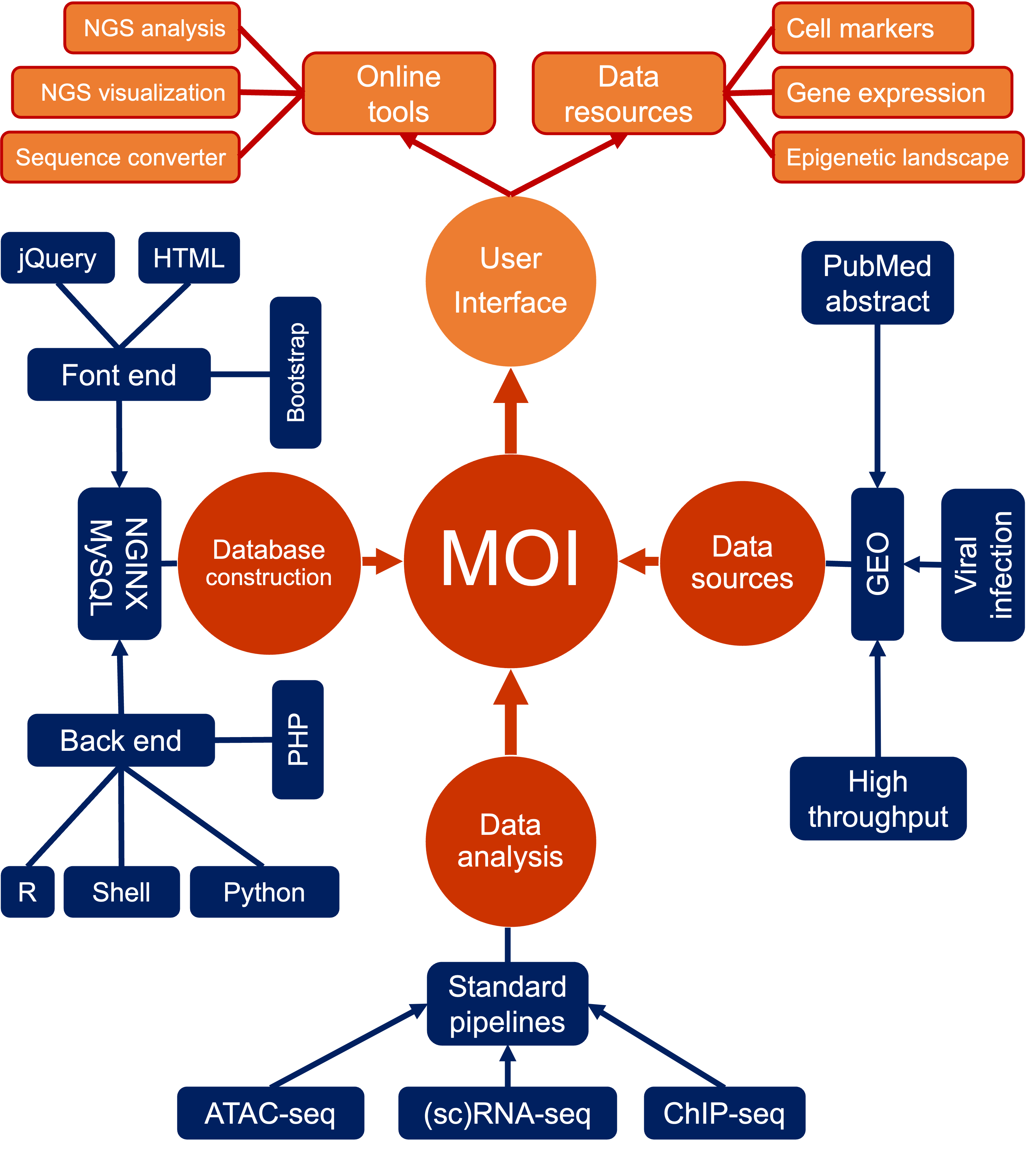
Viruses depend on living cells (host) for replication and proliferation. Viral infections are usually associated with respiratory diseases and cancers. High-throughput sequencing datasets are rapidly accumulating in the field of virology, raising an urgent need to comprehensively collect and efficiently use these data. Here, we developed a Multi-omics Portal of Viral Infection (MOI at http://www.fynn-guo.cn/home.php) in aiming to provide a large number of curated resources on high-throughput sequencing datasets under various virus infections. MOI provides detailed metadata (virus, host, infection time, treatment, assay, etc.) and processed high-dimensional data in host cells.
-
Database content and construct
-
Currently, MOI documents 6,586 manually curated high-throughput sequencing samples from over 77 virus (SARS-CoV-2, SARS-CoV, ZIKV, IAV, etc.) and 33 species (Homo sapiens and Mus musculus, etc.) involving 22 assays (RNA-seq, ChIP-seq, ATAC-seq, BS-seq, CLIP-seq, etc.). For more detailed statistics, please see the Statistics page. Especially, we manually annotated and classified these viruses using the information from ViPR and ICTV databases.
MOI systematically collected multi-omics datasets with virus infection from NCBI/GEO database. Information of viral infections were standardized, including virus, host, assay, infection time, treatment, species, etc. MOI analyzed these datasets using standard pipelines, and provided various results including gene expression, ChIP-seq peaks and annotation, differentially expressed genes, GO-term and KEGG pathway enrichment, alternative splicing events, meta-virus signatures, and etc. MOI integrates customized genome browser and developed multiple tools to help visualize those molecular maps of hosts w/o viral infections.
-
How To Use
-
Search
To query on virus-host interactions, MOI supports three different searching modes: By virus taxonomy, By host and Advanced. Brief search results are presented as a table in the result page. As an example, users can choose Virus Family: Orthomyxviridae, Virus Genus: Betainfluenzavirus, and Virus Name: Influenza B virus in By virus taxonomy mode. Search result are displayed as below, including Experiment ID, Virus Name, Logogram, Host, Assay, Target, Species, GEO ID and PubMed ID. Users can click the "Experiment ID" to view details about each omics dataset in results from search.
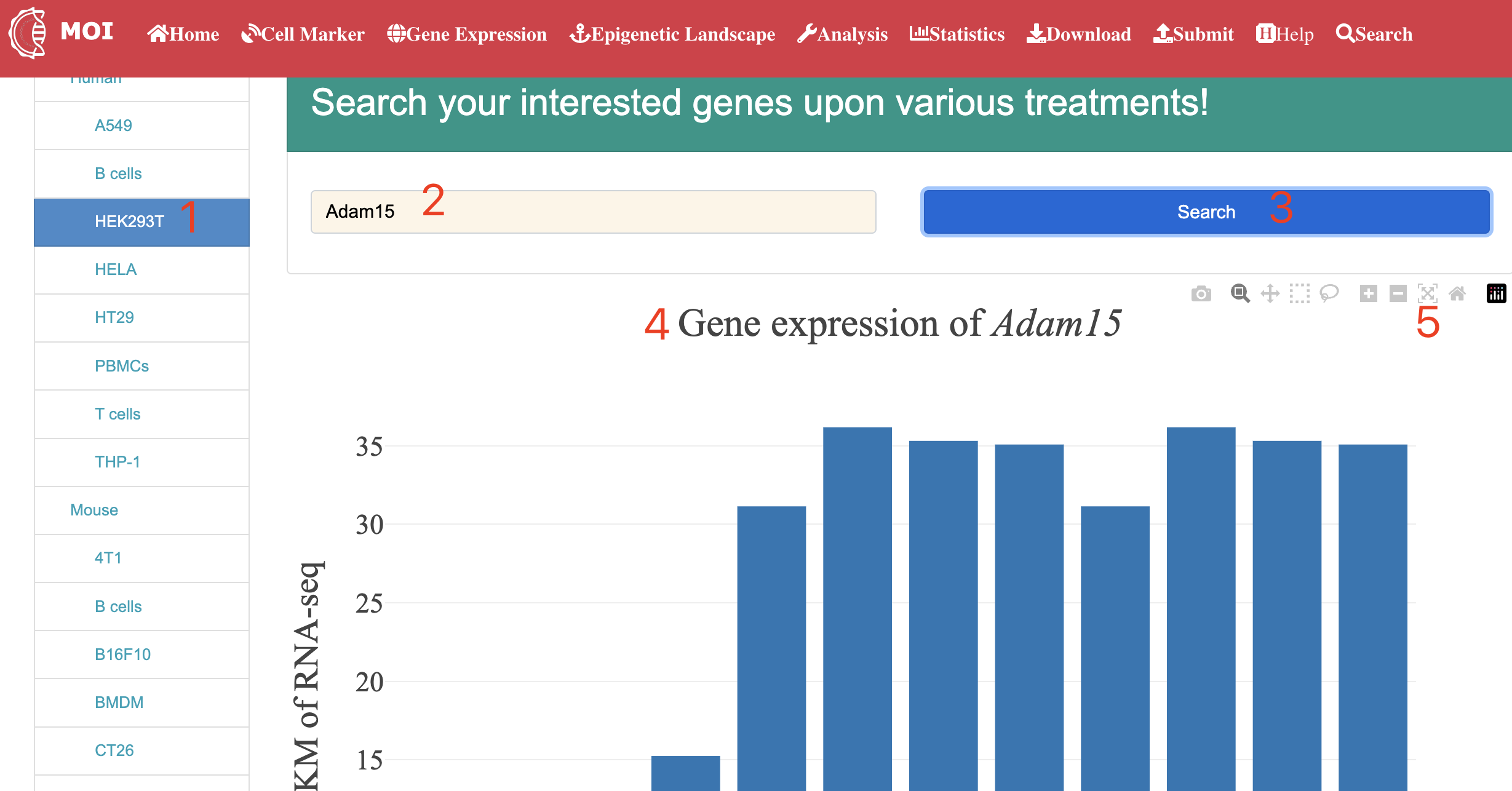
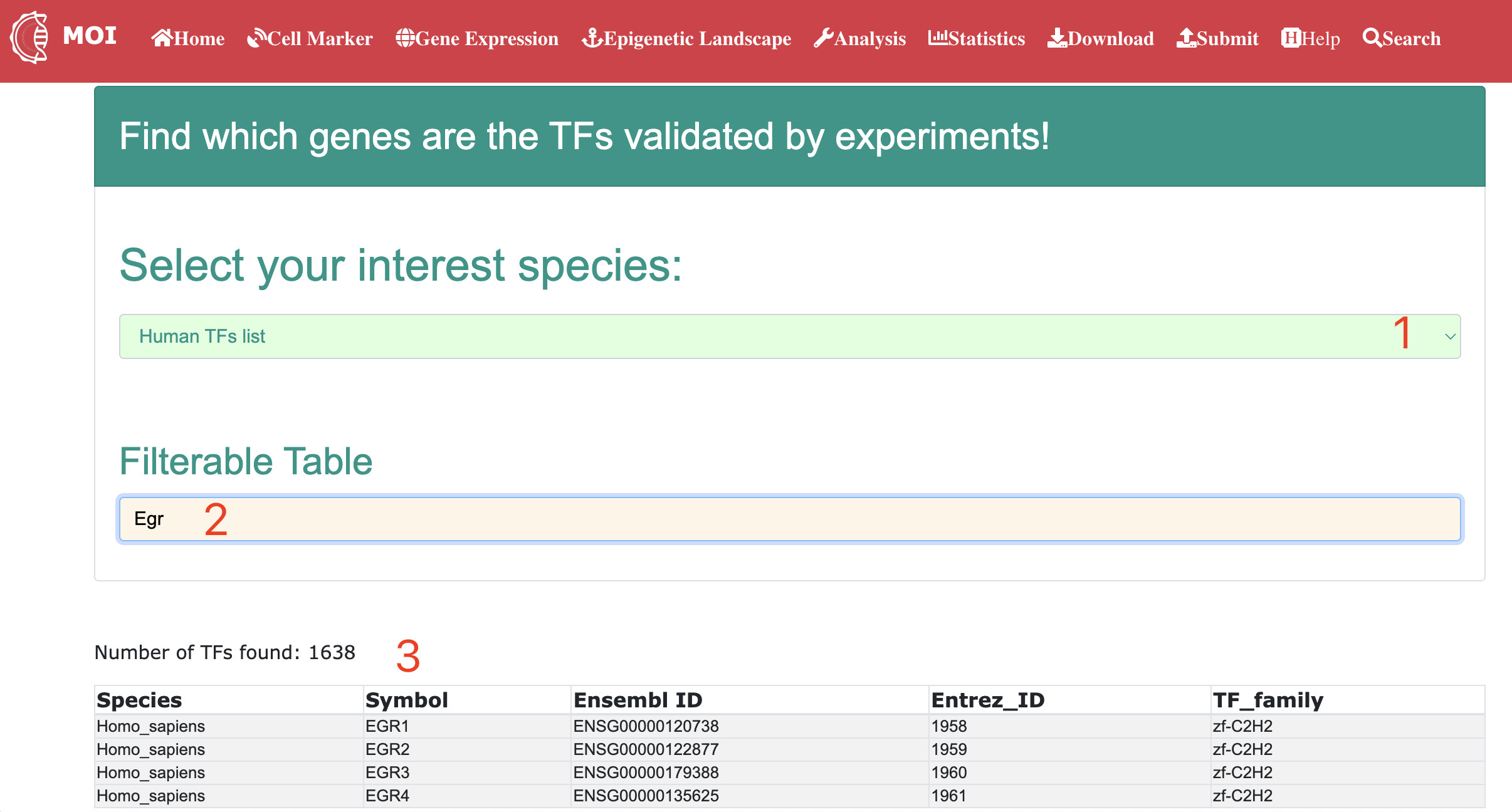
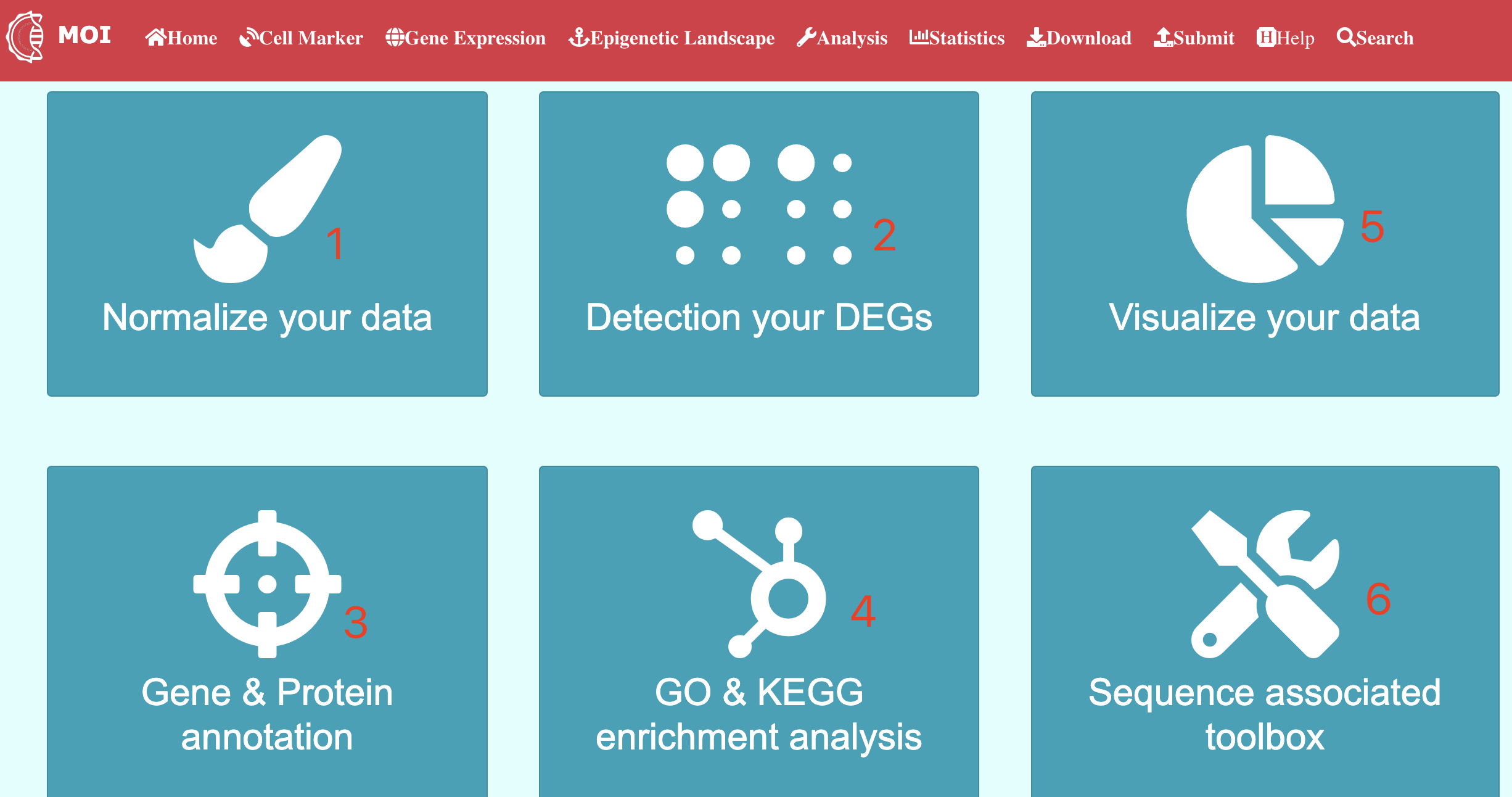
Analysis
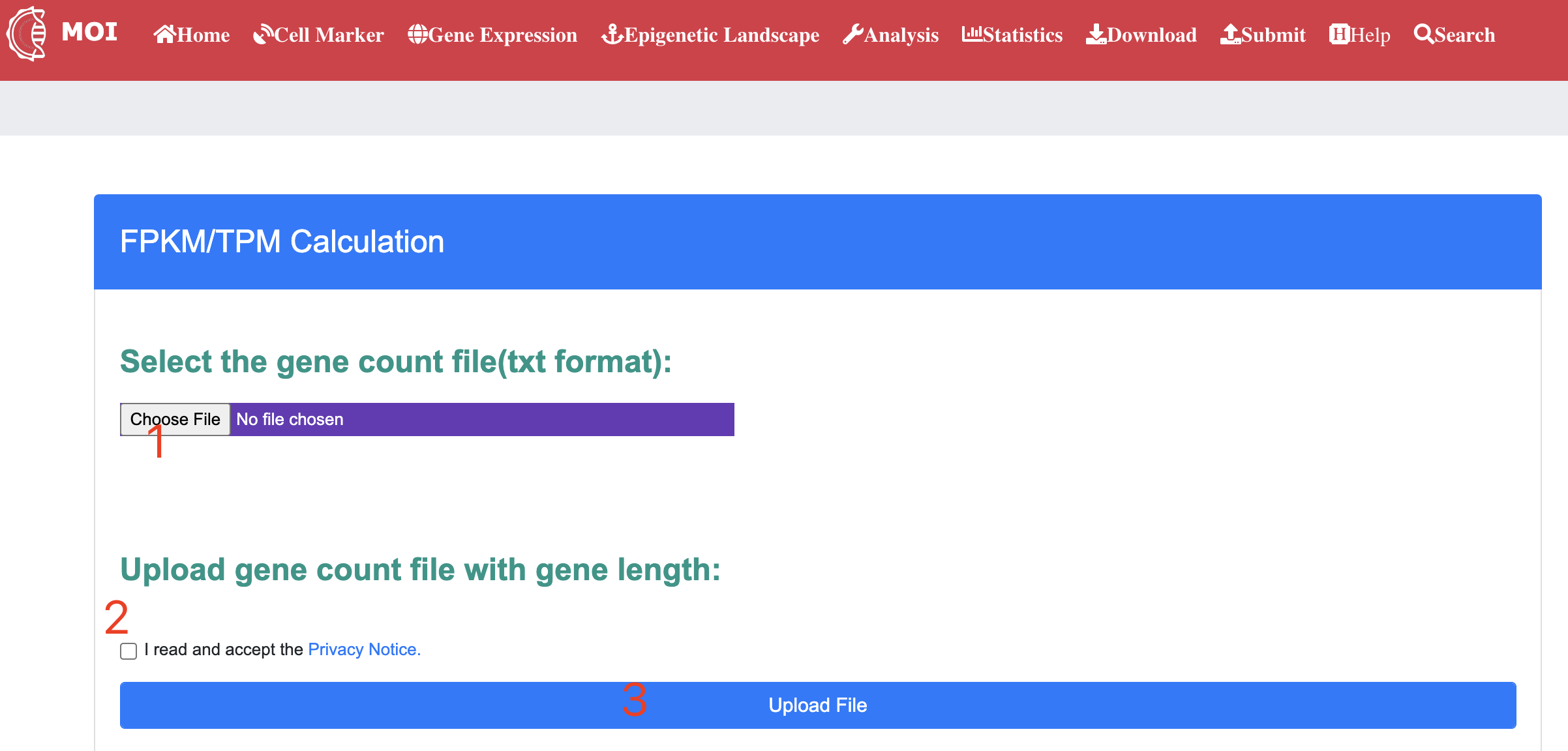
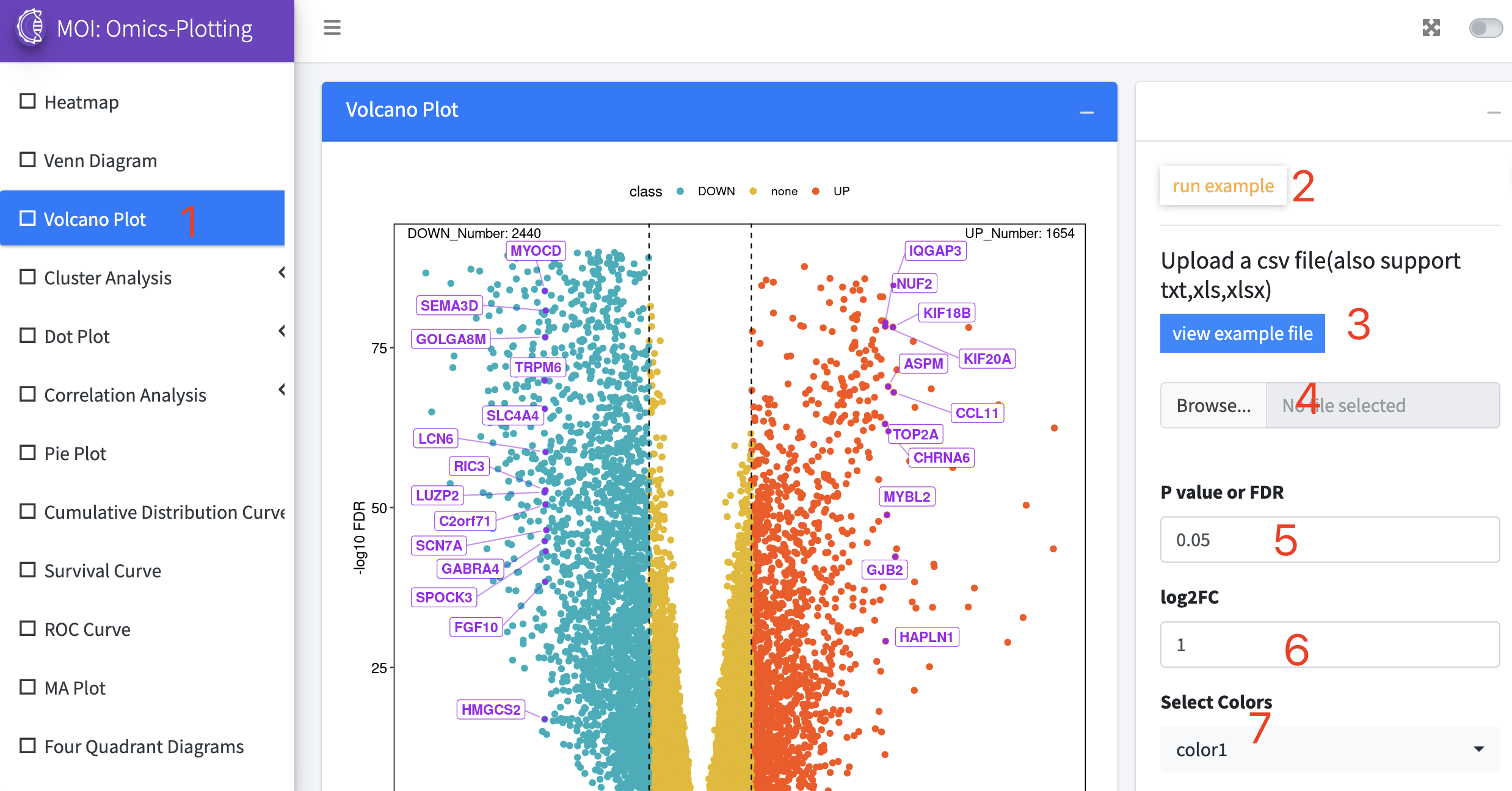
Data analysis and visualization of the multi-scale information in MOI. MOI provides several different analysis tools: 1) "Analyze virus-host interactions", 2) "Analyze dynamic expression profiles by gene", 3) "Analyze expression changes by gene", 4) "Analyze meta-virus signature", 5) "Analyze gene dynamics in time-course assay" and 6) "Analyze scRNA-seq expression".

Track
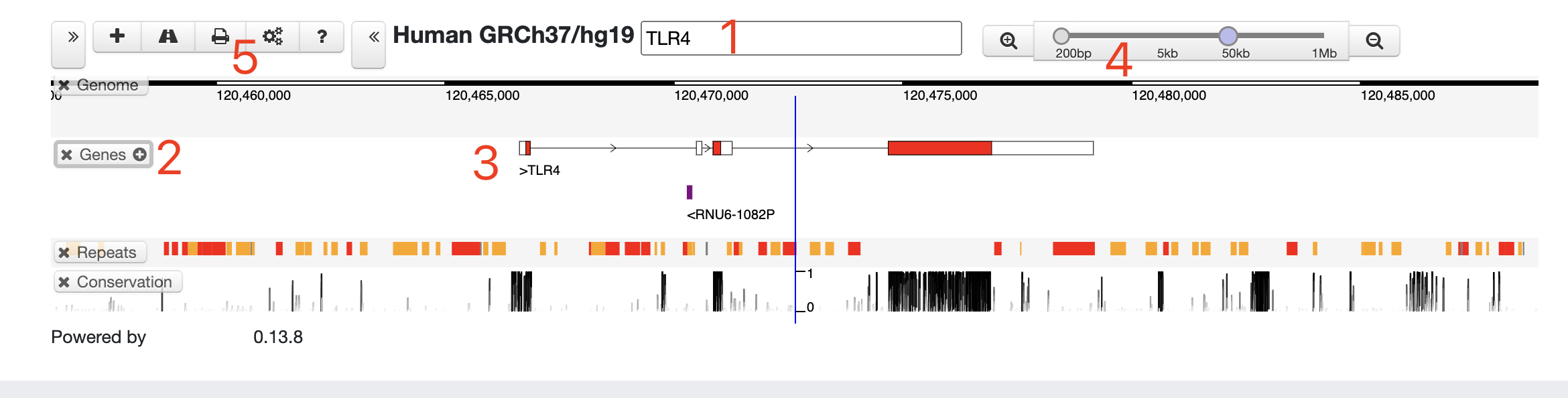
We constructed a MOI Track Hub, allowing visualization of MOI data in UCSC genome browser. The tracks are organized by organism with super-tracks and composite-tracks. The URL for MOI track hub is [http://www.fynn-guo.cn/home.php/db/hub.txt], with which users can connect to add a hub using the http://genome.ucsc.edu/cgi-bin/hgHubConnect#unlistedHubs Then, users can explore the MOI data simultaneously with other existing UCSC data tracks. An example of viewing MOI data in UCSC genome browser is as below.Link
Genome
Integrative genome browser for processed multi-omics data. To help users view the omics-data under various viral infections, we integrated these comprehensive data into MOI, and made a customized genome browser to visualize them simultaneously. These data can also be viewed on UCSC genome browser via connecting MOI track hub.
-
Frequently Asked Questions
-
Why "no results available" is shown for some entries?
Answer: Briefly, the explanation is due to no matched control RNA-seq data for some viral entries, or due to host species having no GO/KEGG annotation or no enriched terms found for some other entries.
Why some virus datasets have no corresponding analysis?
Answer: There are no summarized analysis results beyond mapping for certain types of omics data, such as single cell RNA-seq (scRNA-seq) data due to sample heterogeneity. We plan to process and analyze these omics data in future updates. Meanwhile, we will upgrade the post-mapping analyses such as peak calling when better programs are available.
How to view MOI omic-data through UCSC genome browser?
Answer: We have constructed a MOI track hub (link as above), which can be connected in UCSC genome browser. To know more about UCSC genome browser and track hub, users can following the links to read the corresponding help pages:
http://genome.ucsc.edu/FAQ/FAQlink.html
http://genome.ucsc.edu/goldenPath/help/hgTrackHubHelp.html
-
Development environment
-
The MOI website is hosted on a Linux-based NGINX (V1.21.0) web server, accessed at https://www.nginx.com/. Its user interface is meticulously designed with optimization techniques utilizing the Bootstrap (V4.6.1) framework, accessible at https://getbootstrap.com/docs/4.6/getting-started/introduction/. To facilitate sophisticated data presentation, we have integrated plugins for prominent JavaScript libraries, such as jQuery (V3.6.1), incorporating DataTables (V1.10.19) (https://datatables.net), Biodalliance (v0.13.8) (http://www.biodalliance.org/about.html), and morris.js (v0.5.0) (https://morrisjs.github.io/morris.js/index.html). In the server-side architecture, we deploy PHP (V7.4.24) (http://www.php.net), Python (V3.9.7) (https://www.python.org/), and R (V4.1.3) (https://www.r-project.org/). The MOI database is securely housed within a MySQL (V8.0.24) (http://www.mysql.com) database infrastructure. A meticulous testing regime has been conducted across diverse mainstream web browsers, including Google Chrome, Firefox, Opera, Apple Safari, and Microsoft Edge to ascertain optimal performance and cross-browser compatibility.
The MOI database is freely available for the research community at http://www.fynn-guo.cn/. Users are not required to register or login to access the data in the database.
-
Update
-
MOI was first released on 12-17-2021 and upgraded on March 21, 2021 with more useful on-line analysis tools.
-
Contact us
-
If have any problems or suggestions, please contact Xuefei Guo by e-mail:
